Lab 3: Image Statistics and Data Processing
Introduction
In this lab we will cover the ENVI processing techniques for layer stacking, ,mosaicking and masking datasets. This lab also covers reviewing image statistics and histograms.
Learning Outcomes
- Search for and download Sentinel Data from ESA Copernicus Browser
- Open and evaluate Sentinel-2 data in ENVI
- Perform Layer Stacking operations to combine Sentinel-2 bands
- Mosaicking and color matching datasets
- Creating and Applying Masks
- Evaluating Image Statistics
About the Data
The primary data in this lab is Sentinel-2 Level2A data downloaded from the ESA Copernicus Browser. The goal of this exercise is to download and process Sentinel-2 imagery for Sonoma County, CA for eventual use in image classification. The desired data will be cloud-free, mosaic from June 2025.
Download Sentinel-2 Data and opening in ENVI
- Create a new folder for your lab data, create two subfolders, for original files, and final processed files.
Download the Lab 3 ZIP file from Google Drive or the Z: Drive. The compressed folder contains vector files (shapefile and KMZ approximation) of Sonoma County.
- Go to https://browser.dataspace.copernicus.eu/ and log into your account if you already have and if not click the “Register” button. To register, complete the form and validate your email address to activate your account then log into the Copernicus browser.
- Our goal is to download imagery covering Sonoma County captured earlier this year for eventual use in image classification. To select an area of interest, you can draw a rectangle or polygon or upload an existing file. For the upload, only limited file types are supported, KML/KMZ, GeoJSON, and overly complex, large polygons (many vertices) are also not supported. Upload the simplified Sonoma County KMZ file provided to create an area of interest that represents Sonoma County.
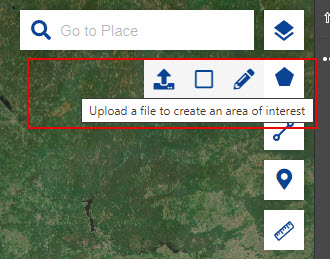
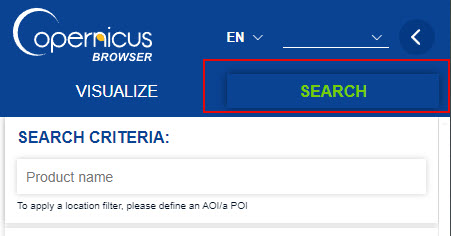
- Click on the Search Tab to search and download Sentinel data.
- Under Data Sources Select Sentinel-2 L2A (Level-2 Surface Reflectance Data) and set the cloud filter to less than 15% or less by dragging the bar below the data source selection.
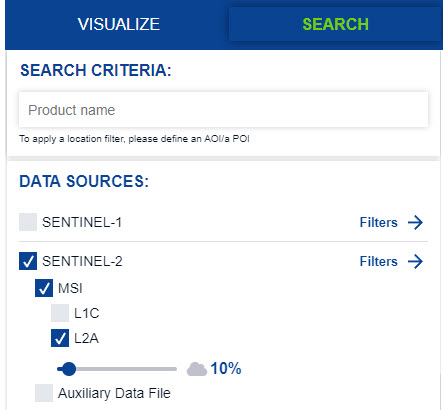
- Scroll down and set the date range for June 15-30, 2025 (leaf-on, growing season imagery) then click the search button.

- There will be many different files that cover the area. Focus on finding two images that cover the majority of Sonoma County and were taken over a close time period (within 2-3 days). Click the Visualize button to see a visual preview of the image. Note the the imagery browser will switch views and you will need to return to the Search tab to download.
- Select the appropriate images and download the files by click the Download Product icon for each associated scene. You will need to download two scenes to cover thr county.
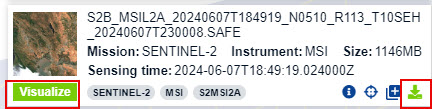
- After the downloads are complete, extract the files into your original data folder, keep the two files in separate folders. Note that you may need to rename the Sentinel-2 ZIP file to a shorter name, as the file names can be too long which causes issues when extracting. When the extracting process is finished you should have a series of folders that contain various data files. Open ENVI.
- In ENVI, select File → Open and navigate to your Originals folder where you extracted your data and find the folder that contains the Sentinel-2 data. Open the metadata XML file that begins with "MTD_MSI...". Only open one of the datasets to start with. After processing the first dataset we will repeat the steps for the second image.
- You should now see the true color Sentinel-2 images. ENVI will automatically display the 10m resolution true color view of the scene and the other bands (20m and 60m) files will be available in the Data Manager. The bands shown in bold are the bands we will use for the analysis.
| Band Number |
Central Wavelength (nm) |
Bandwidth (nm) |
Spatial Resolution (m) |
| B1 |
443 |
20 |
60 |
| B2 |
490 |
65 |
10 |
| B3 |
560 |
35 |
10 |
| B4 |
665 |
30 |
10 |
| B5 |
705 |
15 |
20 |
| B6 |
740 |
15 |
20 |
| B7 |
783 |
20 |
20 |
| B8 |
842 |
115 |
10 |
| B8a |
865 |
20 |
20 |
| B9 |
945 |
20 |
60 |
| B10 |
1375 |
30 |
60 |
| B11 |
1610 |
90 |
20 |
| B12 |
2190 |
180 |
20 |
- Spend some time looking at the different bands available in the data manager. The bands are packaged in "bundles" based on resolution and data type. See the below table for reference. You can select different band combinations and add them to your viewer.
| Bands |
Description |
| 10m-S2MSI |
Surface reflectance for 10m bands (RGB, NIR) |
| 10m-AOT |
Aerosol optical thickness (unitless) |
| 10m-WVP |
Water vapor |
| 10m-TCI |
True color image - an RGB image built from the B02 (Blue), B03 (Green), and B04 (Red) Bands. The reflectance are coded between 1 and 255 |
| 20-SCL |
Scene classification data at 20m resolution |
- Now we will create a Layer Stack to create one raster that includes the first 10 bands of the data in one file (the 10m and 20m resolution surface reflectance data). In the Toolbox select Raster Management → Build Layer Stack. We will also need to subet and mask the data after stacking. We will do this process twice, once for each dataset.

- In the Layer Stack window click to add the layer beside Input Rasters. In the Data Selection Window select the 10 and 20m surface reflectace data associated with the datasets. The names will be 10m-S2MSI2A and 20m-S2MSI2A, hold Ctrl key to select multiple items. Click OK.
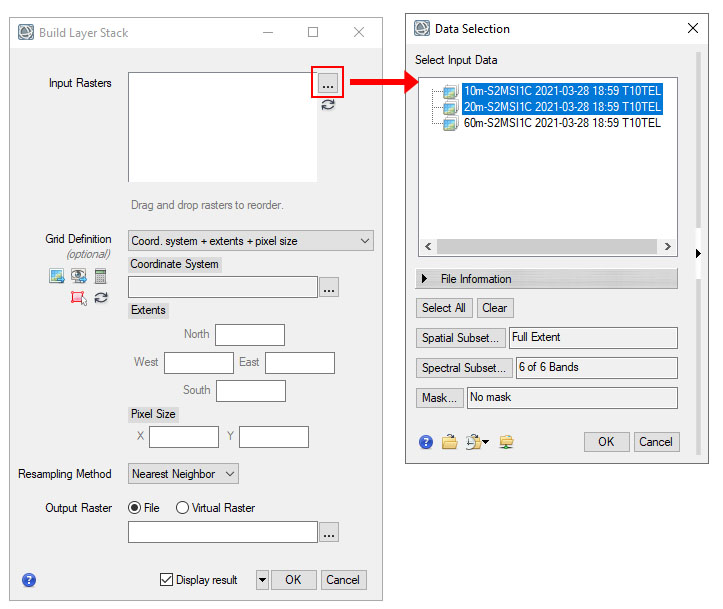
- In the Layer Stack dialog, click the Other tab. This tab allows us to specify the Grid Definition. This includes the coordinate system and pixel size. Under Grid Definition click the From Dataset icon. Select the 20m Sentinel data file as the reference dataset. This will set the coordinate system and cell size based on this dataset.

- Go back to the Main tabin the Layer Stack tool. Check Yes to Order Bands by Wavelength and set the Output Raster to Virtual Raster. The process will create a virtual raster file that will contain the first 10 bands of the Sentinel-2 data ordered from shortest to longest wavelength. Click OK to create and the layer stack. Wait to the image to load and for the pyramid layers to fully load. The stacked image should appear in your viewer when the process is complete. Check to make sure all bands (10) are included. Try making different false color composites to see the different bands to make sure the bands are all visible.
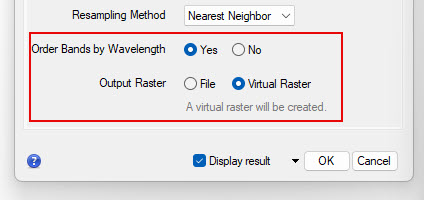
- Now repeat the Layer Stacking process for the other image date, including the same10m and 20m layers in the stack and using the same settings. You should have two stacked virtual rasters when you are completed.
- The last step is to subet and mask the virtual rasters before performing the mosaic operation. Add the Sonoma County shapefile provided to the view. Using the Save As function, subset and mask to the two stacked images. You will need to do this process twice, once for each dataset. Save the rasters as .dat files using the date of the imagery as the filename. Subsetting and masking will reduce the file size and improve the mosaicking process. You may remove the original datasets and virutal rasters from the Data Manager when you are done, leaving only the stacked images.

Mosaic Images
The Seamless Mosaic workflow tool in ENVI allows you to mosaic mutiple georeferenced images into one image. This workflow lets you apply color balancing and edge feathering to create a high-quality mosaic. For detailed instructions and information on the various settings, view Seamless Mosaic in ENVI.
- Now we will mosaic the two datasets together to create one, seamless dataset that can be used for image classification. From the Toolbox launch the Seamless Mosaic Tool. Note that when the tool launches all files will temporarily disappear from the view, don't worry, this is expected and the files are still there!
- Click the + icon and add the two layer stacked images from the previous steps.
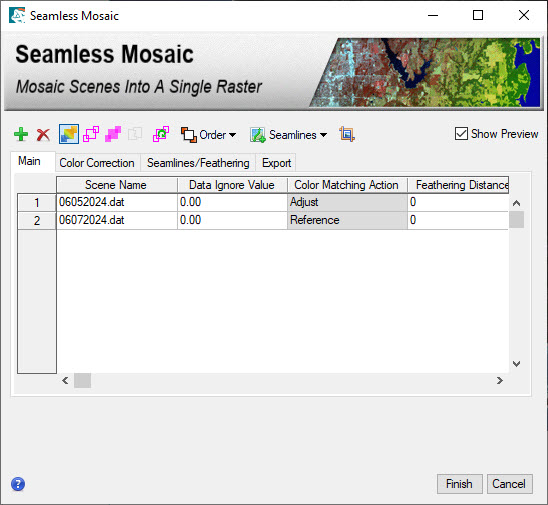
- Click the Seamlines icon and select Auto Generate Seam lines. A seamline is the line along which overlapping scenes will be mosaicked. They help make scene boundaries less visible, ENVI uses an automated geometry-based method to generate seamlines.
- Now click through the various display icons in the mosaic tool to see the different view options. You can turn on and off the foot prints, seamlines and imagery. Check the Show Preview box to see a preview of the mosaic image and turn off all footprints and lines to see a preview of only the final mosaic. . Note the color differences, since they are significant we will use Color Correction techniques.
- Click the Color Correction Tab and check the Histogram Matching box. Try the different options (Overlap Area/Entire Scence) and select the one with the best results. You can also adjust the reference/adjust image in the main tab.
- You can set the feathering distance for one or more scenes, which is the number of pixels across the edge or seamline that scenes will be blended with underlying scenes. Click the Seamlines/Feathering tab and make sure the Seamline Feathering is selected. Click the Main tab, then scroll to the right in the table of input scenes to the Feathering Distance (Pixels) column. Set the feather distance to 10 (10 pixels) for each image.
- Make any final adjustments to the color settings, feathers and seamlines and then click the Export Tab. Keep the default setting (Data Ignore Value should be 0) and save the mosaic file as SonomaCoMosaic.dat in your final folder. The process may take a while.
- Once the mosaic appears in the view, review the image and make sure you are happy with the results before moving forward. The goal is minimize any visual differences between the two images. If there is a clearly visible seamline, you may want to re-do the mosaicing with different settings to minimize the diffrences between the two images.
Edit ENVI Header Metadata
- During the mosaic process, some of the metadata can be lost from the original imagery. we will add this information back to the file by editing the ENVI header file (.hdr) From the Toolbox, select Raster Management > Edit ENVI Header. Select the mosaic raster and click OK.
- Click on the Display Tab and update the Band Names to include a more descriptive name, e.g. NIR or Red. Keep the band number in parentheses, these are the original Sentinel-2 band references. Also update the Description under the main tab to include more information about the dataset - for example what type of data and the dates of the original imagery.
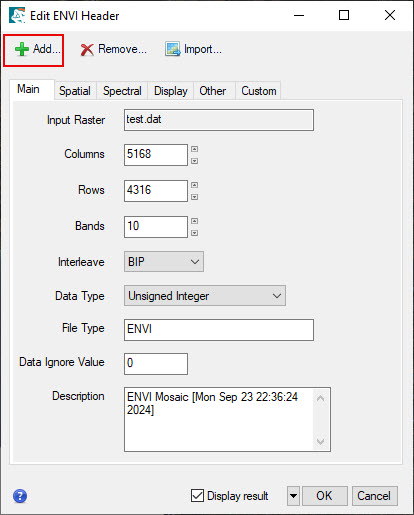
- Click the Add button to select Wavelength and Wavelength Units. Use the information provided in the table at the beginning of the lab instructions to add the correct wavelengths to each of the bands and look up appropriate band names.
- Click OK when you are done and the header file and metadata will be updated. Now the bands names and wavelengths should be visible in the Layer Manager.
View Image Statistics and Histograms
- Now that we have a mosaic with all of the desired bands, we will calculate and view statistics for the image. In the tool box select Statistics → Computer Band Statistics.
- In the Data Selection window select the final Sonoma County Sentinel-2 mosaic you just created and click OK. This will calculate the statistics for all of the bands in the dataset.
- In the Statistics View windows check the Histograms and Covariance boxes. This will add histogram tabls, graphs and covariance statistics to the window.

- Check out the statistics on the screen. The basic stats include the minimum, maximum, mean and standard deviation for each of the bands. Scroll down to see more statistics and detailed distribution statistics for each of the bands. The values for reflectance data should range from ~0-10000, representing 0 to 100% reflectance.
Note that there may be values outside this range that you may want to investigate.
- Below the basic stats are the correlation and covariance tables showing the relationship between bands. All of this same data can also be saved as a text file, so you can review the data without opening ENVI. In the Statistics View select File > Export to Text File. Save the Statistics text file (.txt) in your finals folder. Outside of ENVI, open and review the stats in the text file.
- Now we will view the histograms for all of the bands. In the upper left hand corner click "Select Plot"
 and select "All Histograms". Look at the distribution and shape of the histograms. You can save the histogram by selecting Export → Image. Also check out some of the other plots available. Close the Statistics Windows when you are done. You may need to revisit some of this information to complete the worksheet.
and select "All Histograms". Look at the distribution and shape of the histograms. You can save the histogram by selecting Export → Image. Also check out some of the other plots available. Close the Statistics Windows when you are done. You may need to revisit some of this information to complete the worksheet.
Investigate Low and High Pixel Values
Use the ROI threshold tool or the Raster Color slice tool to investigate various pixel values in the image.
- Create a new ROI on the Sentinel-2 mosaic. Use the ROI threshold tool to find the features in the image with the lowest overall reflectance (across all of the bands) and the highest overall reflectance. Make note of what and where these features are in the image.
- Now try using the raster color slice tool to investigate specific values in the images. Right click on the Sentinel stack layer and select New Raster Color Slice. Select the band you want to use to create a color slice. Specifically locate features that have greater than 100% reflectance in a band.
- Review the worksheet questions. Answer the questions based on the statistics files and the above investigations.
- Back up your final folder. We will need this data for next week's lab!
Turn In - Worksheet and Text file
Upload to Canvas
- Completed Lab 3 Statistics Worksheet
- ENVI generated statistics text file for the Sentinel-2 mosaic(.txt)
- ENVI Header file for the final mosaic
image (.hdr)












